Compute the posterior density of a node value under a fitted Cauchy process on a phylogenetic tree.
ancestral(x, ...)
# S3 method for class 'cauphylm'
ancestral(x, node, values, n_values = 100, n_cores = 1, ...)
# S3 method for class 'cauphyfit'
ancestral(x, node, values, n_values = 100, n_cores = 1, ...)Arguments
- x
- ...
other arguments to be passed to the method.
- node
the vector of nodes for which to compute the posterior density. If not specified, the reconstruction is done on all the nodes.
- values
the vector of values where the density should be computed. If not specified, the reconstruction is done for a grid of
n_valuesvalues between1.5 * min(x$y)and1.5 * max(x$y).- n_values
the number of point for the grid of values. Default to
100. Ignored ifvaluesis provided.- n_cores
number of cores for the parallelization. Default to 1.
Value
an object of S3 class ancestralCauchy,
which is a matrix of posterior values, with nodes in rows and values in columns.
Details
This function assumes a Cauchy Process on the tree with fitted parameters
(see fitCauchy),
and computes the posterior ancestral density of internal nodes,
conditionally on the vector of tip values.
It computes the posterior density on all the points in values,
that should be refined enough to get a good idea of the density curve.
References
Bastide, P. and Didier, G. 2023. The Cauchy Process on Phylogenies: a Tractable Model for Pulsed Evolution. Systematic Biology. doi:10.1093/sysbio/syad053.
See also
Examples
set.seed(1289)
# Simulate tree and data
phy <- ape::rphylo(10, 0.1, 0)
dat <- rTraitCauchy(n = 1, phy = phy, model = "cauchy",
parameters = list(root.value = 10, disp = 0.1))
# Fit the data
fit <- fitCauchy(phy, dat, model = "cauchy", method = "reml")
# Reconstruct the ancestral nodes
anc <- ancestral(fit)
plot_asr(fit, anc = anc, offset = 3)
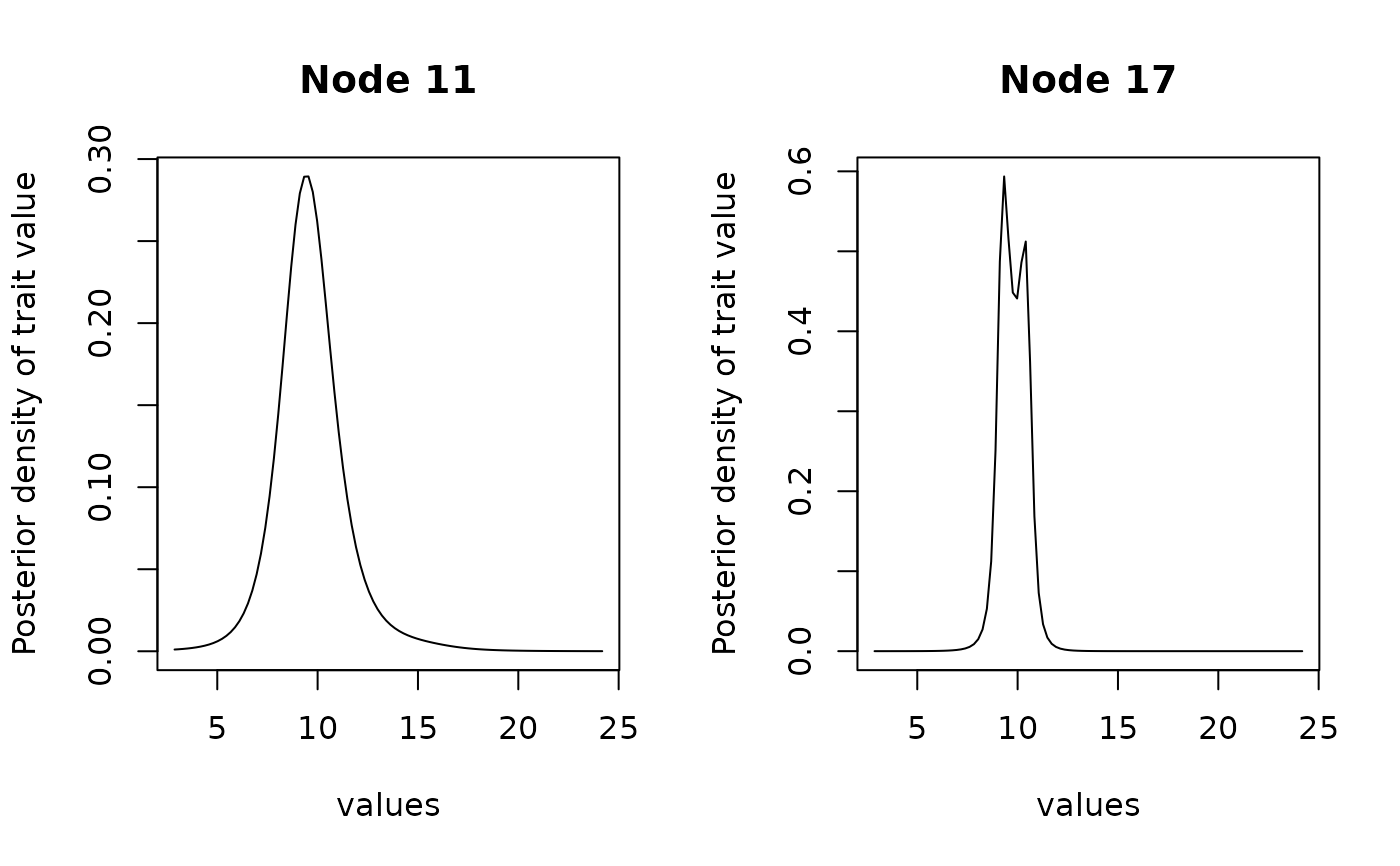
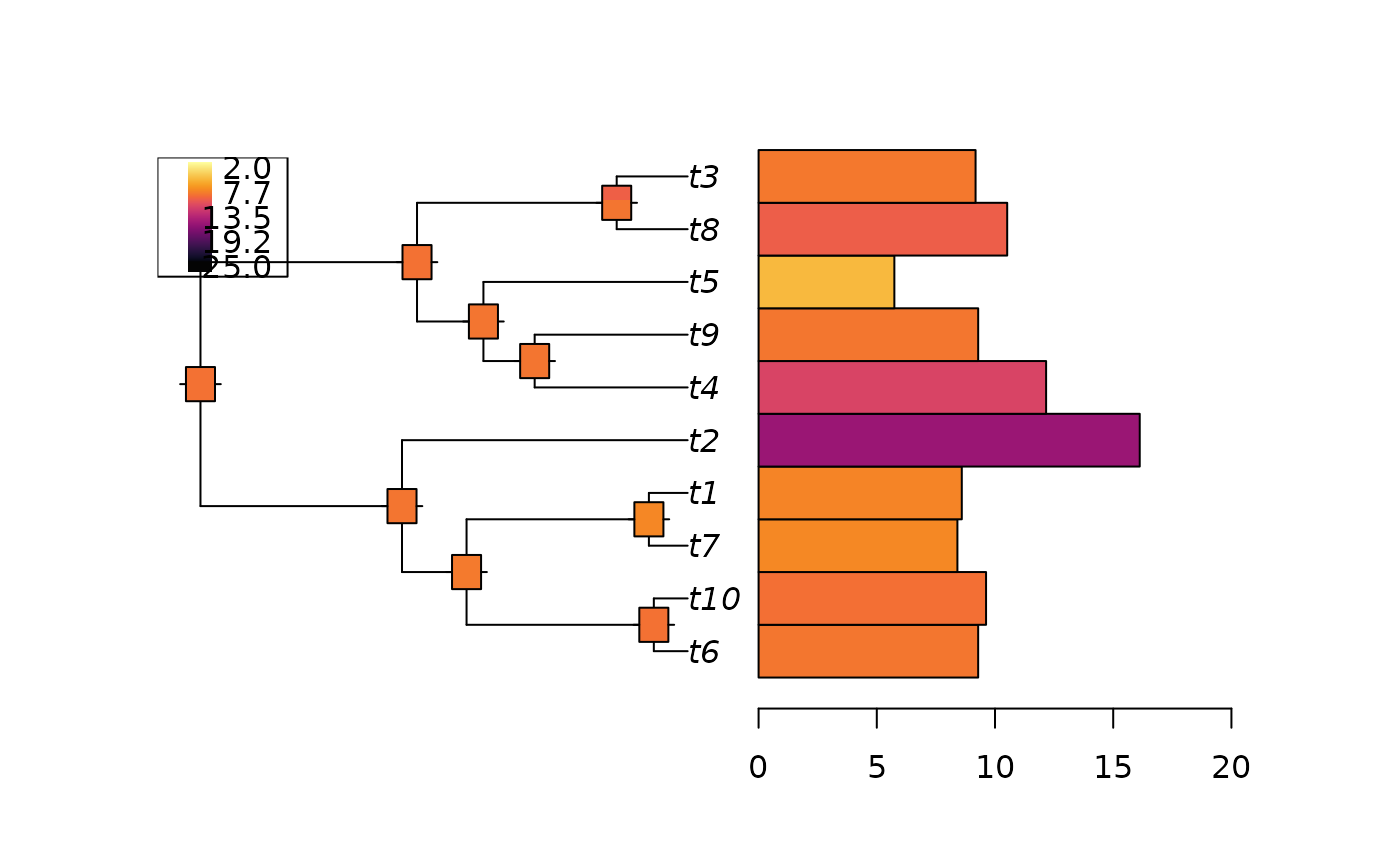 plot(anc, type = "l", node = c(11, 17))
plot(anc, type = "l", node = c(11, 17))
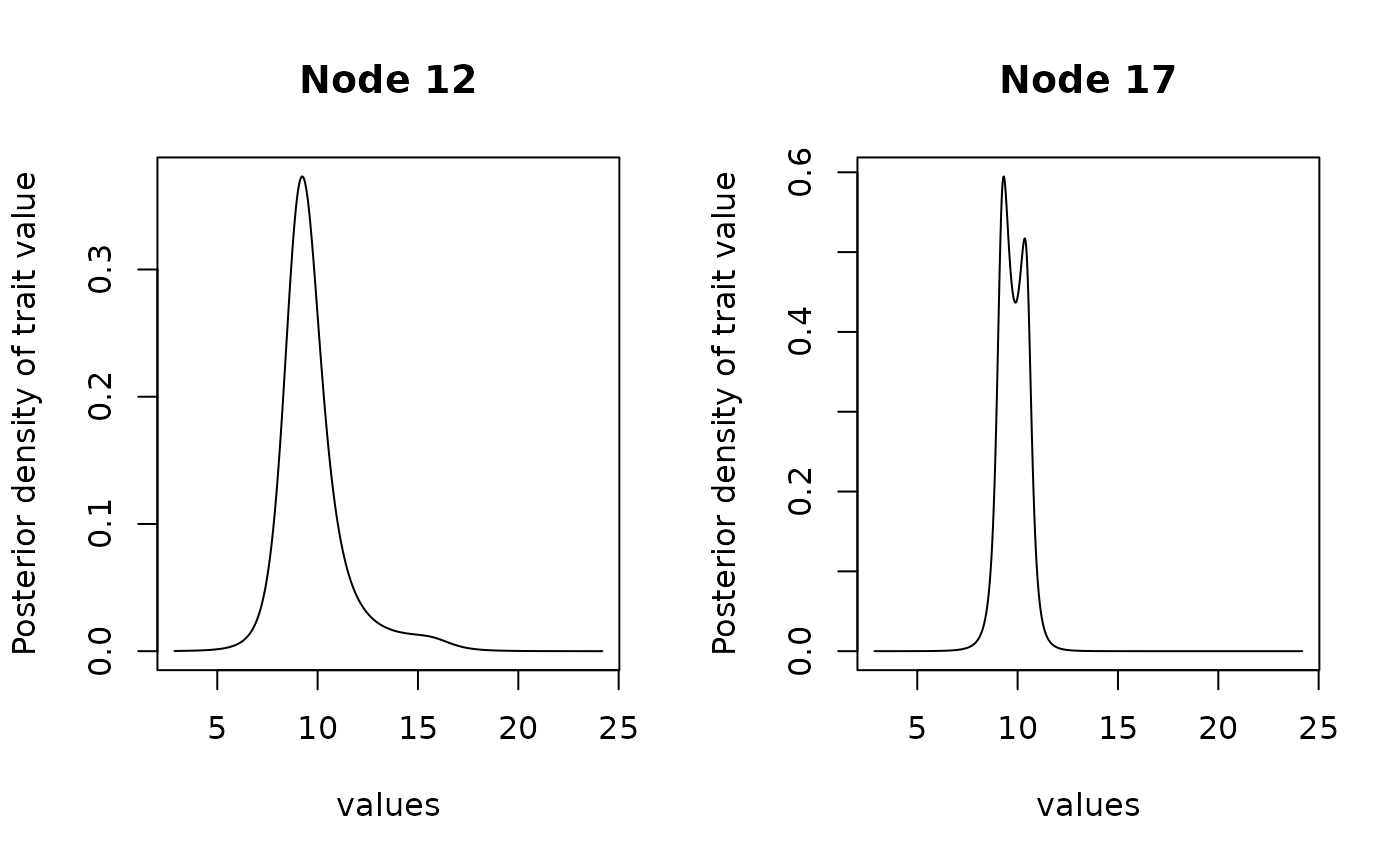
 # Refine grid for node 12 and 17
anc2 <- ancestral(fit, node = c(12, 17), n_values = 1000)
plot(anc2, type = "l")
# Refine grid for node 12 and 17
anc2 <- ancestral(fit, node = c(12, 17), n_values = 1000)
plot(anc2, type = "l")
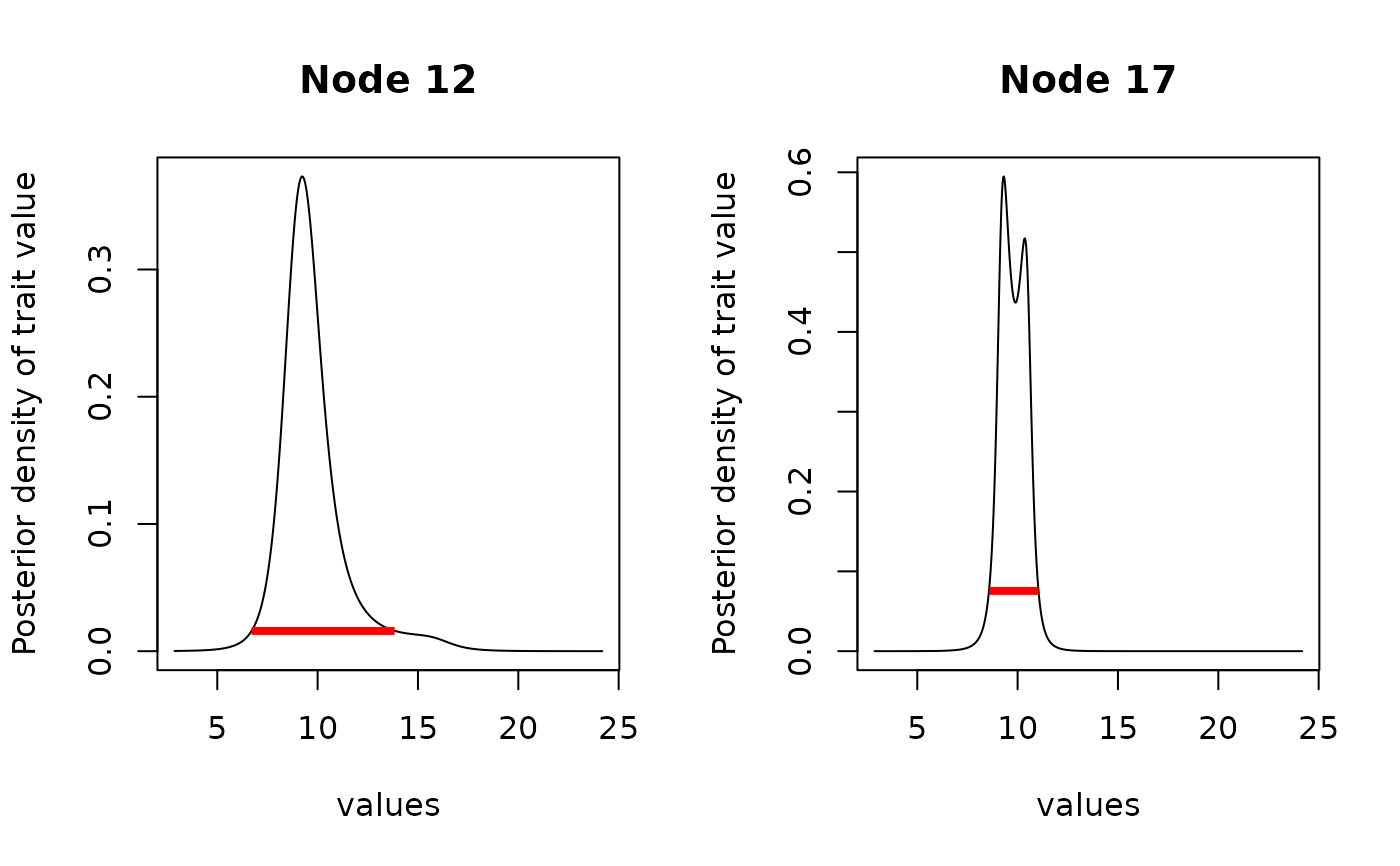
 # Find HDI
library(HDInterval)
hdi_anc <- hdi(anc2)
hdi_anc
#> $`12`
#> begin end
#> [1,] 6.711216 13.83631
#> attr(,"credMass")
#> [1] 0.95
#> attr(,"height")
#> [1] 0.01594822
#>
#> $`17`
#> begin end
#> [1,] 8.588486 11.04174
#> attr(,"credMass")
#> [1] 0.95
#> attr(,"height")
#> [1] 0.07555742
#>
plot(anc2, interval = hdi_anc, type = "l")
# Find HDI
library(HDInterval)
hdi_anc <- hdi(anc2)
hdi_anc
#> $`12`
#> begin end
#> [1,] 6.711216 13.83631
#> attr(,"credMass")
#> [1] 0.95
#> attr(,"height")
#> [1] 0.01594822
#>
#> $`17`
#> begin end
#> [1,] 8.588486 11.04174
#> attr(,"credMass")
#> [1] 0.95
#> attr(,"height")
#> [1] 0.07555742
#>
plot(anc2, interval = hdi_anc, type = "l")